Our research
Current projects involve research in the following areas:
- Norovirus replication and molecular epidemiology
- Adenovirus molecular epidemiology
- Development of antiviral agents against a range of viruses
- Discovering novel viruses in disease reservoirs
- Paleovirology – finding ancient viruses in vertebrate genomes
- Novel viral diagnostic assays and process controls
-
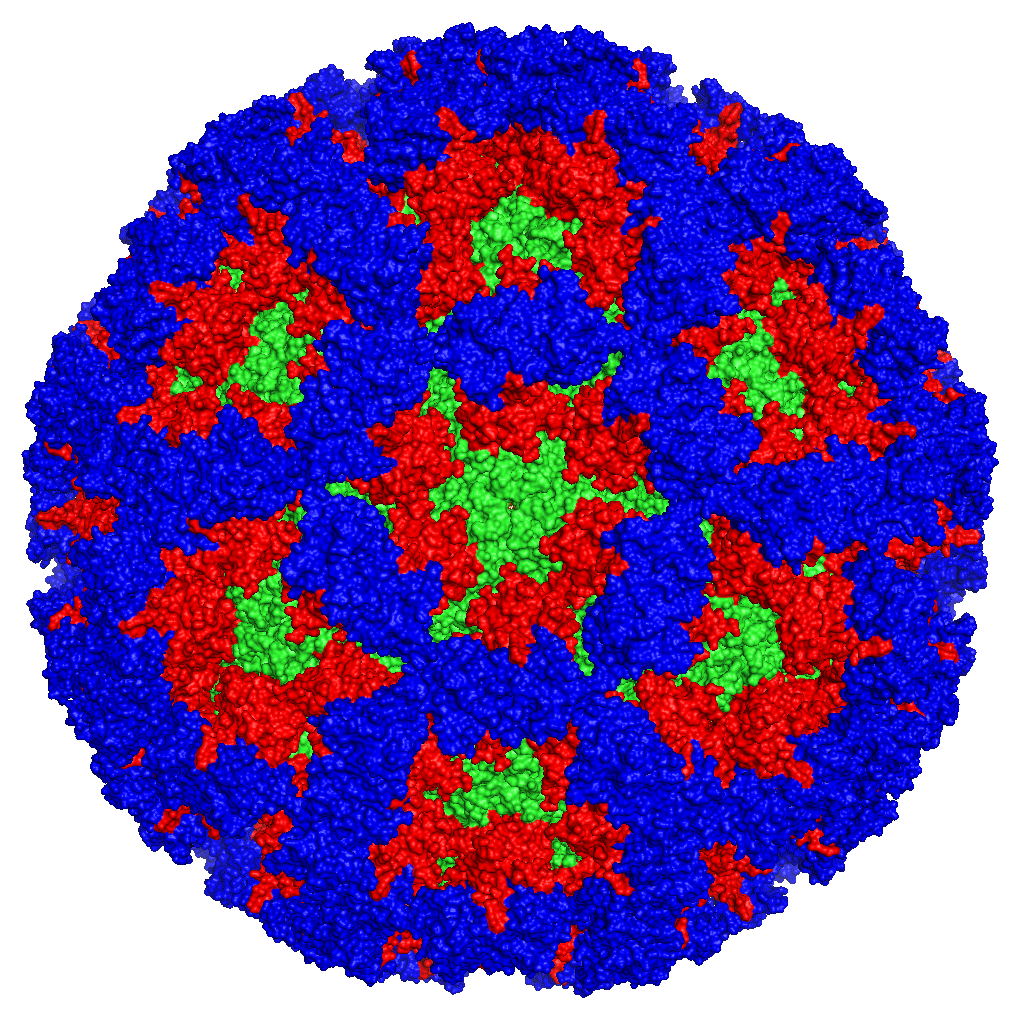
Norovirus (NoV) is now recognised as the leading cause of acute gastroenteritis (AGE) accounting for approximately one fifth of all cases globally. NoV causes illness in around 684 million people every year, resulting in an estimated 218,000 deaths, mainly affecting children within developing countries. Aside from the significant mortality, the global societal costs caused by NoV exceed US $60 billion each year, as a result of health expenditure, morbidity and financial burdens caused by business closures, time off work and hospitalisations. NoV transmission occurs primarily from person-to-person, however, transmission through contaminated food and water makes this virus a significant viral pathogen in terms of foodborne and waterborne outbreaks of gastroenteritis.
Pandemics of acute gastroenteritis are associated with a genetic lineage of noroviruses called GII.4. Peter’s research over the last 18 years has examined evolutionary processes that have led to the emergence of pandemic NoVs. Progress made so far in the molecular biology of NoV by White’s group has revealed the presence of a particularly important genotype of the virus, known as NoV Genogroup II, genotype 4 (GII.4; over 30 other genotypes of NoV exist), as the cause of global pandemics of gastroenteritis, accounting for more than 70% of all NoV infections.
Since the first characterised global pandemic of acute gastroenteritis in the mid-1990s, Peter White's group has been instrumental in identifying and tracing the cause - pandemic GII.4 NoVs. Seminal work by White’s group has shown that the emergence of pandemic GII.4 NoVs is driven by two factors; i) the generation of point mutations in antigenic regions of the viral capsid, in an analogous manner to influenza, and ii) through recombination between two NoVs during a co-infection. Current research from the group on both human and mouse NoV involves development of antiviral agents and research into the host innate immune response, molecular epidemiology, pathogenesis and replication.
-
Human adenoviruses (HAdVs) have been known to infect the gastrointestinal tract in conjunction with the upper or lower respiratory tract and ophthalmologic tissue. These icosahedral, dsDNA viruses can also infect other tissues including neurological tissue. In this study we argue against the current dogma and hypothesise that adenoviral gastroenteritis is not limited to typically ‘enteric’ species F (types 40 and 41). We have recently identified HAdV-A, B, and C. Types associated with clinical gastroenteritis. Interestingly, unlike HAdV-F, these Types are more commonly attributed to tropism in respiratory tissues. Therefore, our findings implicate these Types with undiagnosed acute gastroenteritis.
Using clinical and wastewater samples we aim to build on this hypothesis using cutting edge DNA amplification methods coupled with 3rd generation sequencing techniques and bioinformatics. We will be able to determine the prevalent HAdV species the Sydney and Melbourne populations covering around 3 million people. We also aim to find which unidentified types are associated with acute gastroenteritis in Sydney.
-
Our research focusses on the development of small compound antivirals, including non-nucleoside polymerase inhibitors and nucleoside analogues for the treatment of positive sense RNA viruses, including norovirus, HCV, Zika virus, dengue virus, and other caliciviruses such as feline calicivirus and rabbit haemorrhagic disease virus. These positive sense RNA genomes replicate in the cytoplasm of the infected cell via minus-strand RNA intermediates. In the field of virology, there is an extremely active hunt for new antiviral agents to treat and prevent viral infections.
One target for drug development is the viral RNA dependent RNA polymerase (RdRp) because of its key role in replication. Using our established methodology we have produced highly purified, soluble and active recombinant RdRps from a range of viruses, using Escherichia coli expression systems. High throughput screening (HTS) is a standard platform used to identify lead chemical compounds for drug development. The aim of the antiviral program is to conduct HTS campaigns against the viral RNA polymerases to identify and characterise lead compounds and derivatives for potential antiviral therapy.
-
In 2002, a metagenomics approach – the non-targeted sequencing of all DNA in a sample – was first used to find novel DNA viruses in a marine environment. More recently, such techniques were also developed for the discovery of RNA viruses. For the first time, all viruses in a sample could be identified by their sequence, without the need for extensive culturing or PCR/RT-PCR techniques. Since then, the advent of next generation sequencing (NGS) technologies have greatly facilitated this metagenomic approach to viral discovery. In contrast to Sanger sequencing-based studies, NGS allows the sequencing of millions of base pairs from a sample in a single run, massively increasing the number of viral genomes that can be discovered. NGS has thus revolutionised the field of viral discovery.
The increase in readily-available computational power also permits rapid processing of this data, which can be compared to sequence databases, with, for example, the Basic Local Alignment Search Tool (BLAST) to identify the sample’s taxonomic constituents which for us are RNA viruses. These new techniques have revealed many divergent viral lineages are emerging many of which could pose challenges to the human population. Less than 1% of the earth’s virosphere is estimated to be known suggesting there are millions of potential human pathogens we know nothing about.
-
The study of ancient viruses is termed paleovirology. The aim of this project is to find ancient viruses, or ‘fossil remnants of viruses’. The genomes of animals and insects contain traces of past viral infections through the integration of viral genetic material into the host germline, termed endogenous viral elements (EVEs). These viral fossils can be used to study viruses that existed thousands of years ago. Around 8% of the human genome is comprised of EVEs, of which the vast majority are retroviruses that naturally insert their genomes into the host genome as part of their replication cycle.
For other viruses, germ line integration is rare, but has been documented in many organisms. Using bioinformatics, our lab discovers EVEs in diverse groups of animals. Using genomes from mosquitoes, flies, and ticks, marsupials, including the koala and the Tasmanian Devil, we have identified hundreds of new EVEs, some estimated to be from viruses circulating >100 million years ago. In addition, we have identified unique patterns that link EVEs to small RNA innate immune pathways in both the blacklegged tick Ixodes scapularis and in the Koala. We aim to find more viral fossils in the genomes of other animals, including monetremes which are ecologically threatened, and determine if they are used as a modern day viral defence.
Get in touch
To express your interest in hearing about upcoming projects, please contact:
